Research
Characterizing Uncultivable “Microbial Dark Matter”
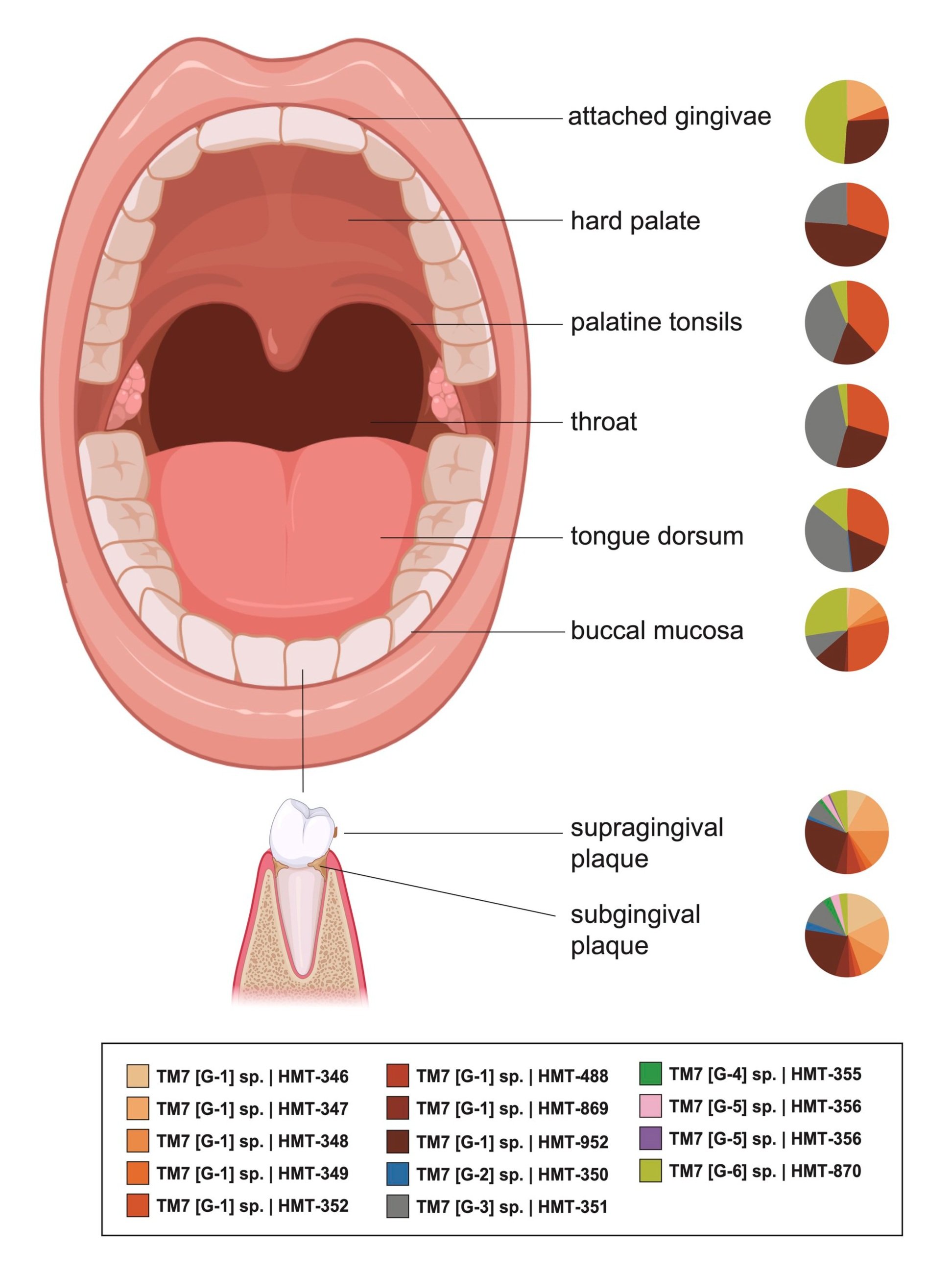
Over 700 species of indigenous bacteria have been identified in the human oral cavity, and approximately one third of these bacteria are not cultivated. The bio-physiological potential hidden among the uncultured bacteria is enormous, and uncovering the commensal and pathogenic potential of yet-to-be cultivated human-associated microbes is crucial to understanding our “microbial organ” and may lead to new cures. One such group of bacteria, the Saccharibacteria (TM7) phylum, has been referred to as “microbial dark matter” due to its lack of cultivable representatives. Our group and collaborators co-isolated the first Saccharibacteria (strain TM7x) together with its bacterial host Schaalia odontolyticus XH001 from the human oral cavity. TM7x lives on the surface of its bacterial hosts and has ultrasmall cell size (200-300 nm) and reduced genome content. Based on our initial studies on TM7x and XH001, we developed a method to culture Saccharibacteria and are able to isolate many more of these bacteria on their host bacteria. We plan to keep refining these methods while also developing new strategies to culture additional Saccharibacteria and their close relatives. There is a clear need for host-microbiome studies to move the field from association to causation-based studies. With the cultivated Saccharibacteria strains and their hosts, we are in a unique position to accomplish this goal and establish the link between these bacteria and mucosal diseases.
Bacteria-Bacteria Interspecies-Interaction
The Candidate Phyla Radiation (CPR) is a major bacterial lineage that comprises one-quarter of all bacterial diversity (>73 phyla) and is found in many different environments, including animals and humans. Despite its ubiquity, Saccharibacteria is the only CPR member with cultivated species in the laboratory and only one of three CPR phyla that has evolved to live in humans; the others are Absconditabacteria (SR1) and Gracilibacteria (GN02). Similar to Saccharibacteria, CPR lineages all share small cell sizes and a reduced genome that is lacking multiple biosynthetic pathways. Based on these properties, most CPR bacteria are predicted to have a symbiotic lifestyle.
Our initial findings demonstrated that Saccharibacteria is an episymbiont that grows obligately on the surface of its host Actinobacteria. It can kill or stably live with its host bacteria, and to date, they cannot be cultured independently. Although various types of interspecies interactions have been reported in bacteria, an obligate episymbiotic interaction is completely novel and there is a great need to understand these highly diverse bacteria. Furthermore, although metagenomic studies established CPR as a ubiquitous member of many microbial communities, their role in these communities is far from clear. Using physiology, molecular cloning, bacterial genetic techniques, and bioinformatic analysis, we are planning to uncover complex interactions between CPR-organisms and their host bacteria.
Saccharibacteria and Human Inflammatory Diseases
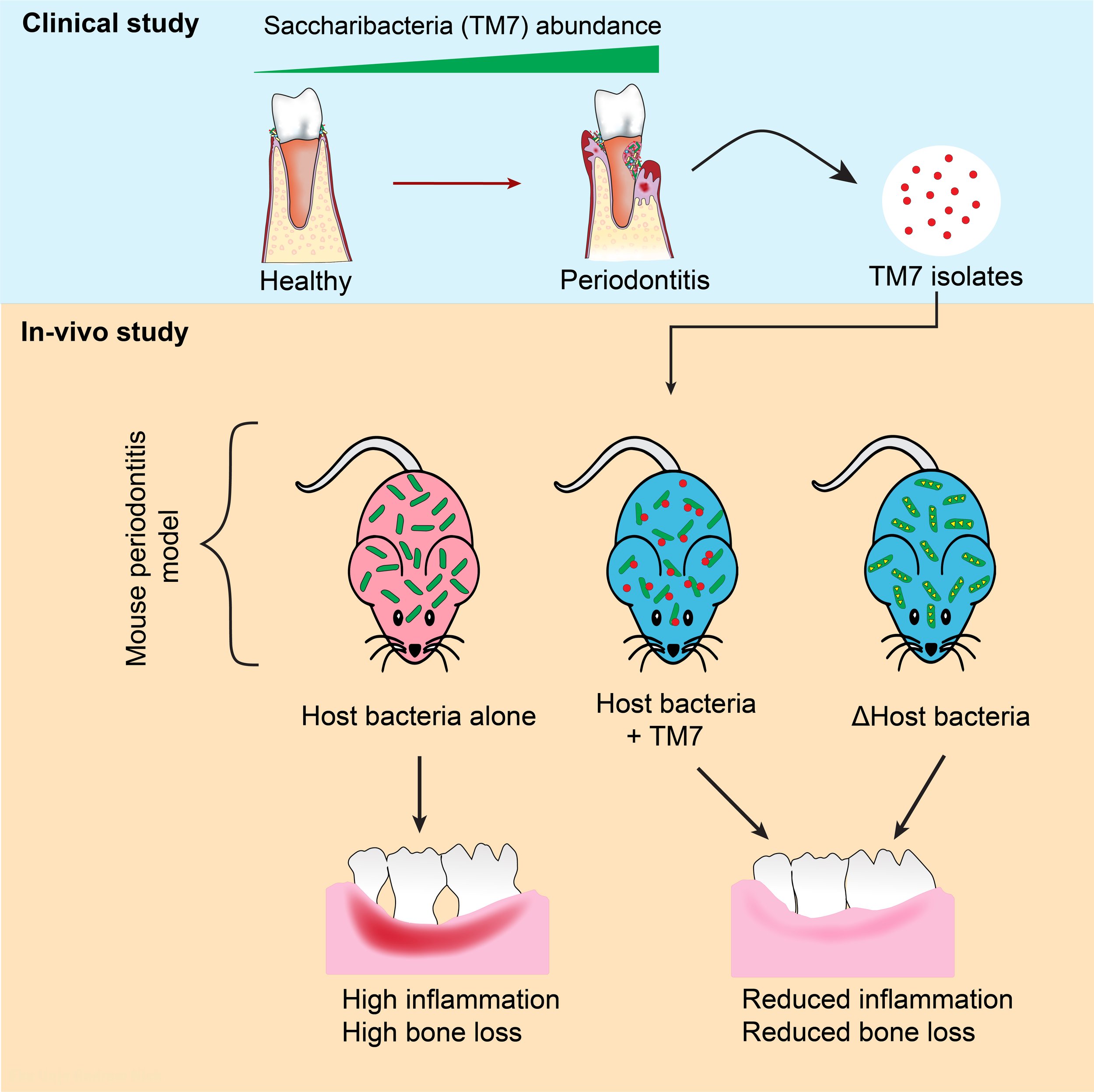
Comprehensive 16S rRNA sequencing studies reported increased relative abundance of Saccharibacteria in plaque samples of patients with chronic/aggressive periodontal disease, comprising more than 20% of the total oral bacterial population in some instances. Furthermore, Saccharibacteria has been linked to various other oral diseases such as gingivitis, halitosis, and peri-implantitis. Elsewhere in the body, they are linked to vaginosis, preterm birth, pelvic inflammatory disease, cervical cancer, inflammatory bowel disease and gastric carcinogenesis. Based on these associations, Saccharibacteria is believed to be one of the pathogenic bacteria involved in the initiation or progression of inflammatory diseases such as periodontitis.
To test this hypothesis, we isolated and cultured multiple Saccharibacteria species from periodontal patients and tested their pathogenic activity. To our surprise, Saccharibacteria reduced inflammatory response and consequential bone loss. This activity occurred through modulation of host bacterial pathogenicity. Going forward, we are applying different tools such as human tissue culture and mouse periodontitis model to investigate the molecular mechanisms behind Saccharibacterial immune suppression and it’s interaction with the eukaryotic host.
CRISPR-Cas13 Based Oral Pathogen Diagnostics
Next-generation sequencing and novel imaging techniques have revealed the complex ways in which microbial pathogens cause oral disease. Now, more than ever, scientists understand the mechanisms and architectures that enable microbes to degrade enamel and promote inflammation. Given the high cost and time of sequencing, however, profiling an individual’s microbiome remains impractical. The field of dental care continues to rely on mechanical intervention rather than microbial modulation. There is an unmet need to translate our increased understanding of oral pathogens from the research bench to the clinic. To address this gap, we are engineering the newly pioneered Cas13 detection platform into a tool capable of identifying oral pathogens directly at the dentist chair within minutes.
Given a saliva sample, Cas13 combined with isothermal amplification can detect DNA within the single molecule range. Unlike current methods of detection, the Cas13 system does not require complex time-consuming procedures, specialized equipment, or technical analysis. Applying this technology to oral care has the potential to shift dentistry away from drilling and filling towards addressing the root cause of oral disease: a patient’s specific microbiome.
Oral Microbiome as Source for Human Systemic Infections
Within the human body, Klebsiella species exhibit a range of roles, serving as colonizing commensals, opportunistic pathogens, or true pathogens, underscoring the considerable adaptability and danger posed by Klebsiella. Among these, K. pneumoniae stands out as the primary culprit behind nosocomial (hospital-acquired) respiratory tract infections and infections in premature intensive care cases, and second most common cause of nosocomial bacterial infections such as bacteremia and urinary tract infections. Notably, oral Klebsiella species can serve as a reservoir for various upper respiratory, gastrointestinal, and nosocomial infections. Despite the wealth of evidence connecting oral Klebsiella to human diseases, there exists a notable gap in our comprehension of how Klebsiella interacts with the normal oral microbiome, particularly concerning colonization ecology, evolution, and resistance. Our current endeavor involves delineating the commensal ecology of Klebsiella species and elucidating their molecular interactions with members of the oral microbiome to decipher their colonization duration and dynamics.